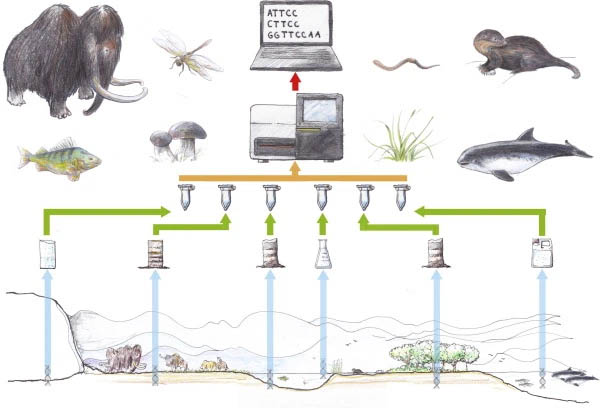
The monitoring and conservation of aquatic populations using conventional methods such as catching are believed harmful to the target and untargeted species. Based on that risk, the researcher examines to develop a new methodology that has fewer impacts and also has a sensitive and accurate result. Since methods in biotechnology-improved rapidly, the opportunity for developing new approaches use biotechnology-based tools is wide open. Several tools that have hi-sensitivity and hi-spectrum possible to be used to create a sophisticated methodology. Interact with organisms; the environment will give us DNA track from saliva, sperm, secretions, eggs, feces, urine, blood, root, leaves, fruit, pollen, etc. Used advanced molecular biology tools, this DNA track can be valuable for more advanced procedures as monitoring and conservation tools.
Environmental DNA (eDNA) is a powerful tool for getting information about species, populations, and behavior with highly sensitive and less destructive than the conventional method. Because of the sensitivity, this technique is usually used for detecting a rare species or invasive species that has a meager population in the environment. Moreover, this methodology is also possible to identify all organism which is living in a varied ecosystem using “DNA barcoding technology.”
The eDNA sample is isolated from environmental samples (water, seawater, soil, or air) and used in several various analyses such as conventional PCR, qPCR, and Genome Sequencer (GS). However, the amount of research showed that the sampling method is the most important step to figure the real population condition in the ecosystem. A lot of aspects such as organic and inorganic compounds, Uv, water flow, depth, pH, organism behavior, and more parameters are believed to influence accuracy. In this case, the sampling methods depend on the environment condition and need to be optimized to get a constant and accurate result.
Several molecular biology analysis can be adapted to analyze eDNA. This review will discuss the strength and weaknesses of each technique.
eDNA – Conventional PCR analysis
Conventional PCR is the cheapest and fastest tool to detect some species which live in the environment. Using a specific primer that can amplify a unique DNA region, this device possibly detects targeted-species. However, this method is not robust to discover rare species with a very deficient population. So, this technique is useful to be used for pre-elementary observation.
eDNA – qPCR analysis
Since a real-time PCR has been developed, detection of trace DNA and quantitative analysis is possible to do. Based on the same reaction with conventional PCR, qPCR machine likely to measure a specific DNA region quantitatively. Syber-green which is dsDNA staining can be used as stepping tools in qPCR. However, this dye is nonspecific that possibly bound to the secondary products. In progress, scientists developed more advanced technology such as the TaqMan probe which is more specific than Sybr-green. This technology will reduce data noise from unspecified PCR amplification products.
Using this superior tool, the discovery of eDNA of rare species is possible without disturb the species and also have super sensitive limits exposure. Moreover, quantitative analysis can even learn the migration behavior and mating season accurately. For example; comparing mitochondrial and genomic DNA (gDNA) is proved as an accurate tool to identify mating season. Sperm that releases in the mating season will increase the concentration of genomic DNA compared to mitochondrial DNA. The population also can be distinguished by gDNA measurement so that migration behavior can be learned using this method.
eDNA – Genome Sequencing (GS) analysis
GS is the most sophisticated machine in molecular biology analysis. This device can read the whole DNA sequence in eDNA sample. Combined with the DNA barcoding database, GS analysis potentially uses to analyze the diversity of the ecosystem and also to classify the organism member of the targeted environment directly from the environmental sample. The result is not only getting qualitative data but also quantitative data.
Other factors in eDNA analysis
However, the result of eDNA analysis does not only depend on the molecular analysis but also depends on the sampling-statistical technique. For example, in the several water eDNA analysis research that showed the water flow, sampling time, and sampling place remarkably influence the detection result and the value of quantitative data. Several factors such as UV, pH, temperature, organo-chemical, and others are possible to affect the results because they can trigger DNA degradation, inhibited the PCR reaction, and also sample homogeneity. In this case, multidisciplinary studies such as hydrology, animal behavior, molecular biology, and others may be needed for designing the perfect experiment.
AQP